Details of the Optogenetic Tool
| General Information of This Photoreceptor (PR) | ||||||
|---|---|---|---|---|---|---|
| PR ID | PR00085 | |||||
| PR Name | Improved Light-Inducible Dimer (iLID) | |||||
| Source of Species | Avena sativa | |||||
| Class | LOV domain | |||||
| UniProt ID | O49003 | |||||
| GenBank | 747155673 | |||||
| PDB ID | 4WF0 | |||||
| Sequence |
LATTLERIEKNFIITDPRLPDNPIIFASDSFLQLTEYSREEILGRNCRFLQGPETDRATV
RKIRDAIDNQTEVTVQLINYTKSGKKFWNVFHLQPMRDYKGDVQYFIGVQLDGTERLHGA AEREAVCLIKKTAFQIAEAANDEYNF |
|||||
| Components | Photoreceptor | Improved Light-Inducible Dimer (iLID) | ||||
| Complementary Protein | Stringent starvation protein B (SspB) | |||||
| Cofactor | Flavin mononucleotide (FMN) | |||||
| 3D Structure | ||||||
| Click to Save PDB File | ||||||
 Cofactor Detail of This PR Cofactor Detail of This PR |
||||||
|---|---|---|---|---|---|---|
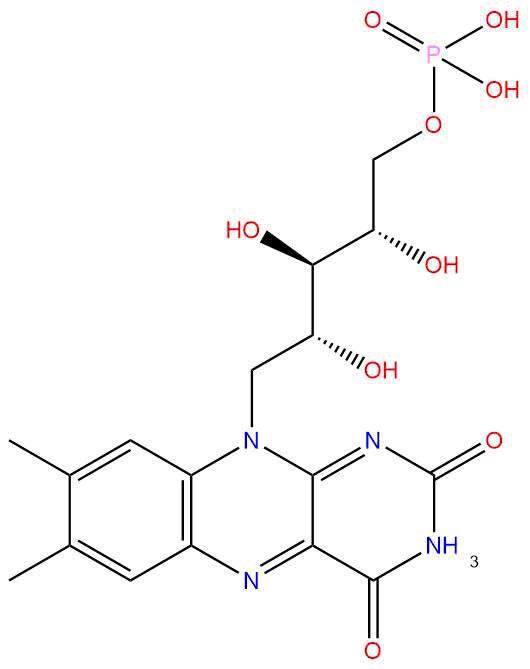
| Cofactor Name | Flavin mononucleotide (FMN) | |||||
| PubChem CID | 643976 | |||||
| Structure |
 |
|||||
| Formula | C17H21N4O9P | |||||
| Molecular Weight | 456.3 | |||||
| Optogenetic System (OS) Consisting of This PR |
|---|
OS Name: Light regulated signal protein localization and activity system 1 (LRPLA 1)
[1]
OS Name: Light regulated signal protein localization and activity system 2 (LRPLA 2)
[1]
OS Name: Light-induceble tethering system (LIT)
[2]
OS Name: Opto-dHTH, an optogenetic inhibitor to completely disrupt GqGTP-PLC interactions 1 (Opto-dHTH 1)
[3]
OS Name: Opto-dHTH, an optogenetic inhibitor to completely disrupt GqGTP-PLC interactions 2 (Opto-dHTH 2)
[3]
OS Name: Opto-dHTH, an optogenetic inhibitor to completely disrupt GqGTP-PLC interactions 3 (Opto-dHTH 3)
[3]
OS Name: Opto-dHTH, an optogenetic inhibitor to completely disrupt GqGTP-PLC interactions 4 (Opto-dHTH 4)
[3]
OS Name: Opto-dHTH, an optogenetic inhibitor to completely disrupt GqGTP-PLC interactions 5 (Opto-dHTH 5)
[3]
OS Name: Optogenetic control Cdc42 system (Opto-Cdc42)
[4]
OS Name: Optogenetic control Exoc70 for membrane traffiking system 2 (Opto-Exoc70-MT 2)
[5]
OS Name: Optogenetic control Rac system (Opto-Rac)
[4]
OS Name: Optogenetic mitochondira deformation system 2 (OptoMD 2)
[8]
OS Name: Optogenetic mitochondira deformation system 3 (OptoMD 3)
[8]
OS Name: Optogenetic recuit factors to microtubule plus ends system 1 (Opto-RFMT 1)
[9]
OS Name: Optogenetic recuit factors to microtubule plus ends system 2 (Opto-RFMT 2)
[9]
OS Name: Optogenetic recuit factors to microtubule plus ends system 3 (Opto-RFMT 3)
[9]
OS Name: Optogenetic recuit factors to microtubule plus ends system 4 (Opto-RFMT 4)
[9]
OS Name: Optogenetic regulate iTrkA activity by iLID system (Opto-iTrkA-iLID)
[10]
OS Name: Optogenetic regulate iTrkB activity by iLID system (Opto-iTrkB-iLID)
[10]
OS Name: Optogenetics of LOV domains with chemiluminescence 2 (Opto-chemilu 2)
[11]
OS Name: Optogentic protein recruitment by iLID system 1 (OptoPR-iLID 1)
[12]
OS Name: Optogentic protein recruitment by iLID system 2 (OptoPR-iLID 2)
[12]
OS Name: Optogentic protein recruitment by iLID system 3 (OptoPR-iLID 3)
[12]
OS Name: Optogentic protein recruitment by iLID system 4 (OptoPR-iLID 4)
[12]
OS Name: Optogentic regulate stromal interaction molecule 1 system 5 (OptoSTIM1-5)
[13]
| References |
|---|
 Detail Info
Detail Info