Details of the Optogenetic Tool
| General Information of This Photoreceptor (PR) | ||||||
|---|---|---|---|---|---|---|
| PR ID | PR00304 | |||||
| PR Name | Residue1-621 of Phytochrome B (PhyB (1-621)) | |||||
| Source of Species | Arabidopsis | |||||
| Class | Phytochromes | |||||
| UniProt ID | P14713 | |||||
| GenBank | NP_179469 | |||||
| PDB ID | 4OUR | |||||
| Sequence |
MVSGVGGSGGGRGGGRGGEEEPSSSHTPNNRRGGEQAQSSGTKSLRPRSNTESMSKAIQQ
YTVDARLHAVFEQSGESGKSFDYSQSLKTTTYGSSVPEQQITAYLSRIQRGGYIQPFGCM IAVDESSFRIIGYSENAREMLGIMPQSVPTLEKPEILAMGTDVRSLFTSSSSILLERAFV AREITLLNPVWIHSKNTGKPFYAILHRIDVGVVIDLEPARTEDPALSIAGAVQSQKLAVR AISQLQALPGGDIKLLCDTVVESVRDLTGYDRVMVYKFHEDEHGEVVAESKRDDLEPYIG LHYPATDIPQASRFLFKQNRVRMIVDCNATPVLVVQDDRLTQSMCLVGSTLRAPHGCHSQ YMANMGSIASLAMAVIINGNEDDGSNVASGRSSMRLWGLVVCHHTSSRCIPFPLRYACEF LMQAFGLQLNMELQLALQMSEKRVLRTQTLLCDMLLRDSPAGIVTQSPSIMDLVKCDGAA FLYHGKYYPLGVAPSEVQIKDVVEWLLANHADSTGLSTDSLGDAGYPGAAALGDAVCGMA VAYITKRDFLFWFRSHTAKEIKWGGAKHHPEDKDDGQRMHPRSSFQAFLEVVKSRSQPWE TAEMDAIHSLQLILRDSFKES |
|||||
| Components | Photoreceptor | Residue1-621 of Phytochrome B (PhyB (1-621)) | ||||
| Complementary Protein | Phytochrome interacting factor 3 (PIF3) | |||||
| Cofactor | Phycocyanobilin (PCB) | |||||
| 3D Structure | ||||||
| Click to Save PDB File | ||||||
 Cofactor Detail of This PR Cofactor Detail of This PR |
||||||
|---|---|---|---|---|---|---|
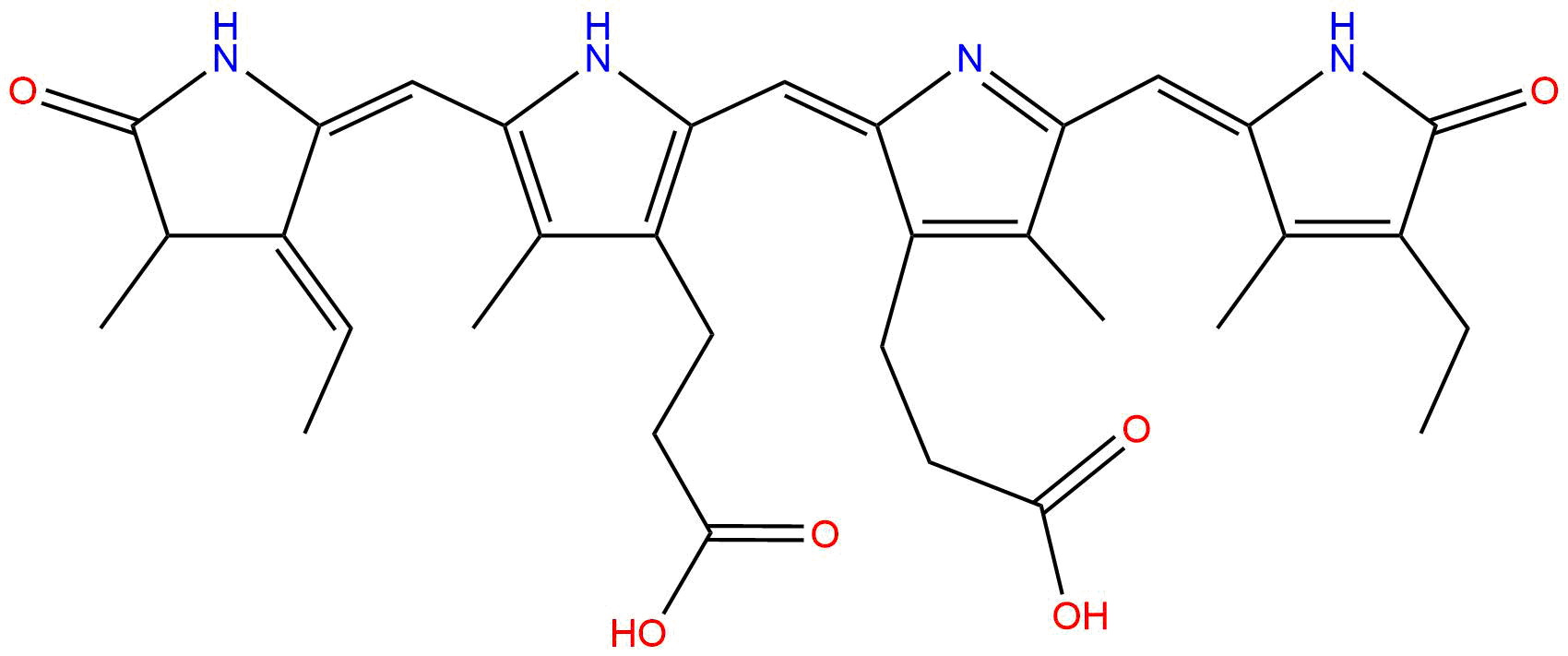
| Cofactor Name | Phycocyanobilin (PCB) | |||||
| PubChem CID | 5280816 | |||||
| Structure |
 |
|||||
| Formula | C33H38N4O6 | |||||
| Molecular Weight | 586.7 | |||||
| Optogenetic System (OS) Consisting of This PR |
|---|
OS Name: Optogenetic control heterologous metabolism system (Opto-HM)
[3]
OS Name: Optogenetic control protein location system based on PhyB (Opto-CPL-PhyB)
[4]
OS Name: Optogenetic control SOS activity system (Opto-SOS)
[5]
OS Name: Optogenetic control transcription activation system 2 (Opto-CTA 2)
[6]
| References |
|---|
 Detail Info
Detail Info