Details of the Optogenetic Tool
| General Information of This Photoreceptor (PR) | ||||||
|---|---|---|---|---|---|---|
| PR ID | PR00367 | |||||
| PR Name | Cryptochromes 2 (1-535; E490G) (CRY2olig (535)) | |||||
| Source of Species | Arabidopsis thaliana | |||||
| Class | Cryptochromes | |||||
| UniProt ID | Q96524 | |||||
| GenBank | NP_849588 | |||||
| PDB ID | 6X24 | |||||
| Mutation Site | E490G | |||||
| Sequence |
MKMDKKTIVWFRRDLRIEDNPALAAAAHEGSVFPVFIWCPEEEGQFYPGRASRWWMKQSL
AHLSQSLKALGSDLTLIKTHNTISAILDCIRVTGATKVVFNHLYDPVSLVRDHTVKEKLV ERGISVQSYNGDLLYEPWEIYCEKGKPFTSFNSYWKKCLDMSIESVMLPPPWRLMPITAA AEAIWACSIEELGLENEAEKPSNALLTRAWSPGWSNADKLLNEFIEKQLIDYAKNSKKVV GNSTSLLSPYLHFGEISVRHVFQCARMKQIIWARDKNSEGEESADLFLRGIGLREYSRYI CFNFPFTHEQSLLSHLRFFPWDADVDKFKAWRQGRTGYPLVDAGMRELWATGWMHNRIRV IVSSFAVKFLLLPWKWGMKYFWDTLLDADLECDILGWQYISGSIPDGHELDRLDNPALQG AKYDPEGEYIRQWLPELARLPTEWIHHPWDAPLTVLKASGVELGTNYAKPIVDIDTAREL LAKAISRTRGAQIMIGAAPDEIVADSFEALGANTIKEPGLCPSVSSNDQQVPSAV |
|||||
| Components | Photoreceptor | Cryptochromes 2 (1-535; E490G) (CRY2olig (535)) | ||||
| Complementary Protein | Cryptochrome-interacting basic-helix-loop-helix 1 (CIB1/CIBN) | |||||
| Cofactor | Flavin adenine dinucleotide (FAD) | |||||
| 3D Structure | ||||||
| Click to Save PDB File | ||||||
 Cofactor Detail of This PR Cofactor Detail of This PR |
||||||
|---|---|---|---|---|---|---|
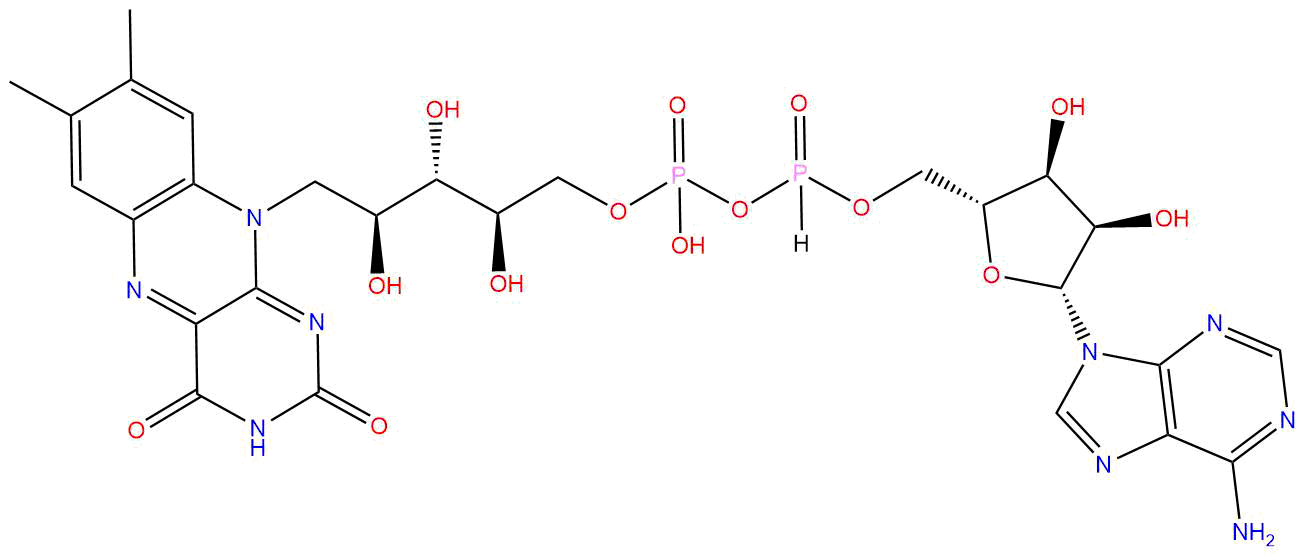
| Cofactor Name | Flavin adenine dinucleotide (FAD) | |||||
| PubChem CID | 643975 | |||||
| Structure |
 |
|||||
| Formula | C27H33N9O15P2 | |||||
| Molecular Weight | 785.5 | |||||
| Optogenetic System (OS) Consisting of This PR |
|---|
OS Name: LARIAT-based approach for synaptic vesicle clustering 2 (LARIAT 2)
[1]
Functional protein: Arabidopsis thalianaCryptochrome-2 variants with C-terminal charge alteration (535)
 Detail Info
Detail Info